Chapter 8. Inferene Engines¶
import os
import warnings
import arviz as az
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import jax.numpy as jnp
from jax import random, vmap, local_device_count, pmap, lax, tree_map, hessian
from jax import nn as jnn
from jax.scipy import stats, special
import numpyro
import numpyro.distributions as dist
import numpyro.optim as optim
from numpyro.infer import MCMC, NUTS, HMC, Predictive
from numpyro.diagnostics import hpdi, print_summary
from numpyro.infer import Predictive, SVI, Trace_ELBO, init_to_value
from numpyro.infer.autoguide import AutoLaplaceApproximation
seed=1234
if "SVG" in os.environ:
%config InlineBackend.figure_formats = ["svg"]
warnings.formatwarning = lambda message, category, *args, **kwargs: "{}: {}\n".format(
category.__name__, message
)
az.style.use("arviz-darkgrid")
numpyro.set_platform("cpu") # or "gpu", "tpu" depending on system
numpyro.set_host_device_count(local_device_count())
# import pymc3 as pm
# import numpy as np
# import pandas as pd
# import scipy.stats as stats
# import matplotlib.pyplot as plt
# import arviz as az
# az.style.use('arviz-darkgrid')
Non-Markovian methods¶
Grid computing¶
def posterior_grid(grid_points=50, heads=6, tails=9):
"""
A grid implementation for the coin-flipping problem
"""
grid = jnp.linspace(0.01, 1, grid_points)
prior = jnp.repeat(1/grid_points, grid_points) # uniform prior
likelihood = jnp.exp(dist.Binomial(total_count=(heads+tails), probs=grid).log_prob(heads))
posterior = likelihood * prior
posterior /= posterior.sum()
return grid, posterior
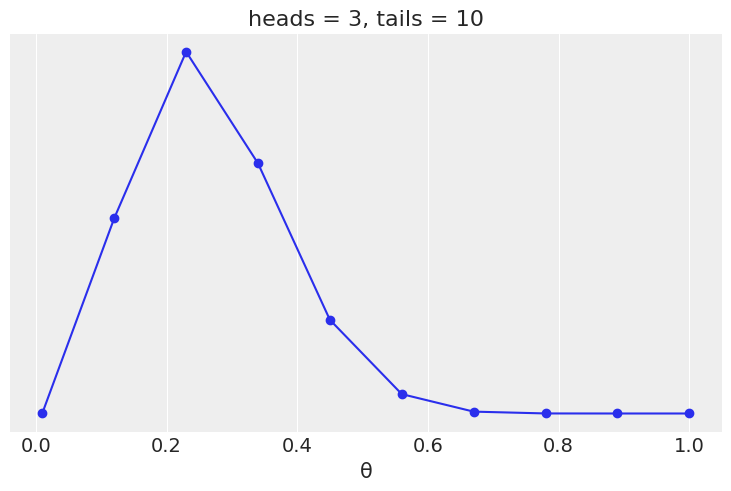
Assuming we flip a coin 13 times and we observed 3 head we have:
data = jnp.repeat(jnp.array([0, 1]), jnp.asarray((10, 3)))
points = 10
h = data.sum()
t = len(data) - h
grid, posterior = posterior_grid(points, h, t)
plt.plot(grid, posterior, 'o-')
plt.title(f'heads = {h}, tails = {t}')
plt.yticks([])
plt.xlabel('θ');
Quadratic method¶
def model(obs=None):
p = numpyro.sample('p', dist.Beta(concentration1=1., concentration0=1.))
w = numpyro.sample('w', dist.Binomial(total_count=1, probs=p), obs=obs)
guide = AutoLaplaceApproximation(model)
svi = SVI(model, guide, optim.Adam(1), Trace_ELBO(), obs=data)
svi_result = svi.run(random.PRNGKey(0), 20000)
params = svi_result.params
params
100%|████████████████████| 20000/20000 [00:01<00:00, 11830.65it/s, init loss: 11.2695, avg. loss [19001-20000]: 8.6997]
{'auto_loc': DeviceArray([-1.0116092], dtype=float32)}
samples = guide.sample_posterior(random.PRNGKey(0), params, (10000,))
xs = jnp.sort(samples['p'])
std_q = jnp.std(xs)
mean_q = jnp.mean(xs)
mean_q, std_q
(DeviceArray(0.28205067, dtype=float32),
DeviceArray(0.11372714, dtype=float32))
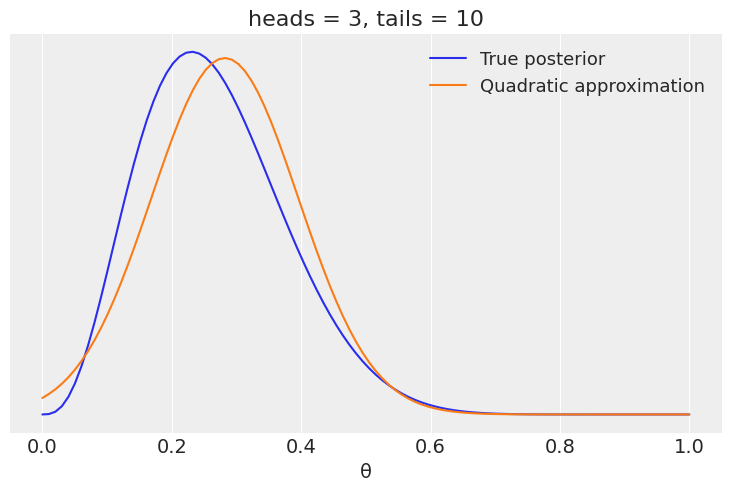
# analytical calculation
x = jnp.linspace(0, 1, 100)
plt.plot(x, jnp.exp(dist.Beta(h+1, t+1).log_prob(x)), label='True posterior')
# quadratic approximation
plt.plot(x, jnp.exp(dist.Normal(mean_q, std_q).log_prob(x)),label='Quadratic approximation')
# plt.plot(x, guide.(params).sample(random.PRNGKey(0), sample_shape=(len(x),)))
plt.legend(loc=0, fontsize=13)
plt.title(f'heads = {h}, tails = {t}')
plt.xlabel('θ', fontsize=14)
plt.yticks([])
([], [])
quantiles = guide.quantiles(params, 0.5)
quantiles
{'p': DeviceArray(0.26666504, dtype=float32)}
guide.median(params)
{'p': DeviceArray(0.26666504, dtype=float32)}
# predictive = Predictive(guide, params=params, num_samples=1000)
# samples = predictive(random.PRNGKey(1), data)
numpyro.diagnostics.hpdi(samples['p'], prob=0.5)
array([0.17510453, 0.32284716], dtype=float32)
# with pm.Model() as normal_aproximation:
# p = pm.Beta('p', 1., 1.)
# w = pm.Binomial('w',n=1, p=p, observed=data)
# mean_q = pm.find_MAP()
# std_q = ((1/pm.find_hessian(mean_q, vars=[p]))**0.5)[0]
# mean_q['p'], std_q
Markovian methods¶
Monte Carlo¶
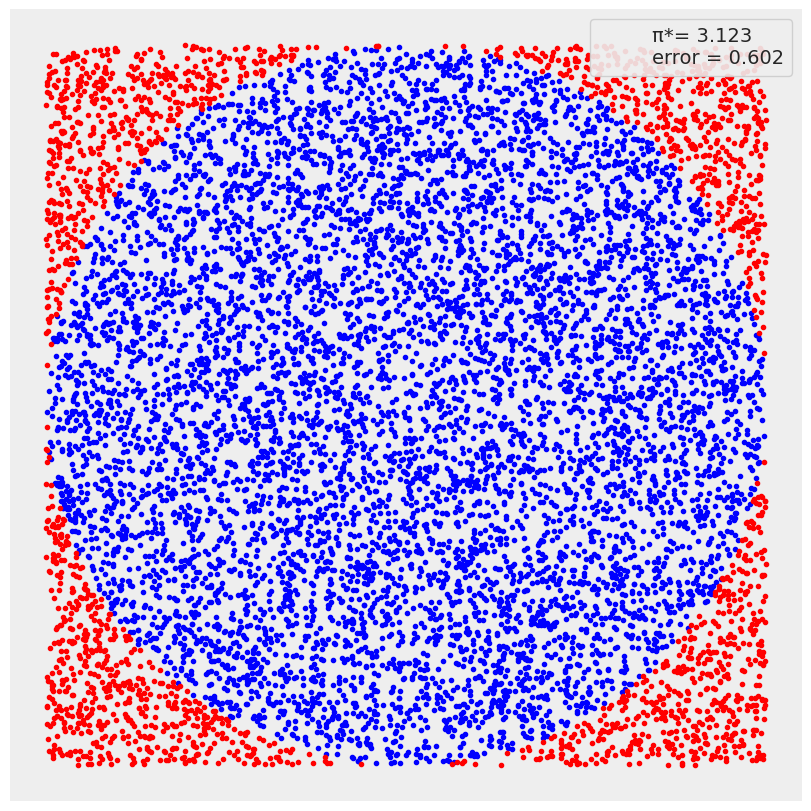
N = 10000
x, y = dist.Uniform(low=-1, high=1).sample(random.PRNGKey(0), sample_shape=(2, N))
inside = (x**2 + y**2) <= 1
pi = inside.sum()*4/N
error = abs((pi - jnp.pi) / pi) * 100
outside = jnp.invert(inside)
plt.figure(figsize=(8, 8))
plt.plot(x[inside], y[inside], 'b.')
plt.plot(x[outside], y[outside], 'r.')
plt.plot(0, 0, label=f'π*= {pi:4.3f}\nerror = {error:4.3f}', alpha=0)
plt.axis('square')
plt.xticks([])
plt.yticks([])
plt.legend(loc=1, frameon=True, framealpha=0.9)
<matplotlib.legend.Legend at 0x7ffb946d5be0>
def metropolis(func, draws=10000):
"""A very simple Metropolis implementation"""
trace = jnp.zeros(draws)
old_x = 0.5 # func.mean()
old_prob = jnp.exp(func.log_prob(old_x))
delta = dist.Normal(loc=0, scale=0.5).sample(random.PRNGKey(0), sample_shape=(draws,))
for i in range(draws):
new_x = old_x + delta[i]
new_prob = jnp.exp(func.log_prob(new_x))
acceptance = new_prob / old_prob
# if acceptance >= dist.Uniform(low=0, high=1).sample(random.PRNGKey(i)):
if acceptance >= random.uniform(random.PRNGKey(i)):
trace = trace.at[i].set(new_x)
old_x = new_x
old_prob = new_prob
else:
# x[idx] = y``, use ``x = x.at[idx].set(y)
trace = trace.at[i].set(old_x)
return trace
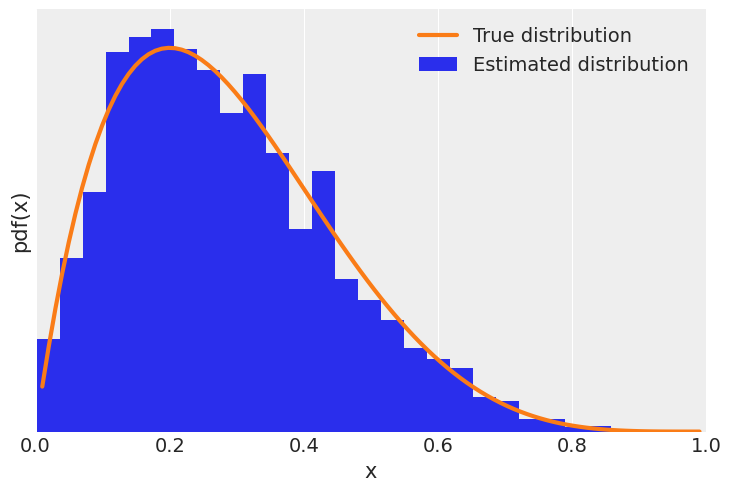
func = dist.Beta(concentration1=2, concentration0=5)
trace = metropolis(func=func)
x = jnp.linspace(0.01, .99, 100)
y = jnp.exp(func.log_prob(x))
plt.xlim(0, 1)
plt.plot(x, y, 'C1-', lw=3, label='True distribution')
plt.hist(trace[trace > 0], bins=25, density=True, label='Estimated distribution')
plt.xlabel('x')
plt.ylabel('pdf(x)')
plt.yticks([])
plt.legend()
<matplotlib.legend.Legend at 0x7ffb7765be50>
Diagnosing the samples¶
def centered_model(obs=None):
a = numpyro.sample('a', dist.HalfNormal(scale=10))
b = numpyro.sample('b', dist.Normal(loc=0, scale=a), sample_shape=(10,))
kernel = NUTS(centered_model, target_accept_prob=0.85)
trace_cm = MCMC(kernel, num_warmup=1000, num_samples=1000, num_chains=2, chain_method='sequential')
trace_cm.run(random.PRNGKey(seed))
def non_centered_model(obs=None):
a = numpyro.sample('a', dist.HalfNormal(scale=10))
b_offset = numpyro.sample('b_offset', dist.Normal(loc=0, scale=1), sample_shape=(10,))
b = numpyro.deterministic('b', 0 + b_offset * a)
kernel = NUTS(non_centered_model, target_accept_prob=0.85)
trace_ncm = MCMC(kernel, num_warmup=1000, num_samples=1000, num_chains=2, chain_method='sequential')
trace_ncm.run(random.PRNGKey(seed))
sample: 100%|██████████████████████████| 2000/2000 [00:02<00:00, 794.15it/s, 31 steps of size 2.11e-01. acc. prob=0.92]
sample: 100%|██████████████████████████| 2000/2000 [00:00<00:00, 6585.73it/s, 7 steps of size 1.84e-01. acc. prob=0.81]
sample: 100%|███████████████████████████| 2000/2000 [00:02<00:00, 858.42it/s, 7 steps of size 3.92e-01. acc. prob=0.92]
sample: 100%|██████████████████████████| 2000/2000 [00:00<00:00, 7252.10it/s, 7 steps of size 5.03e-01. acc. prob=0.89]
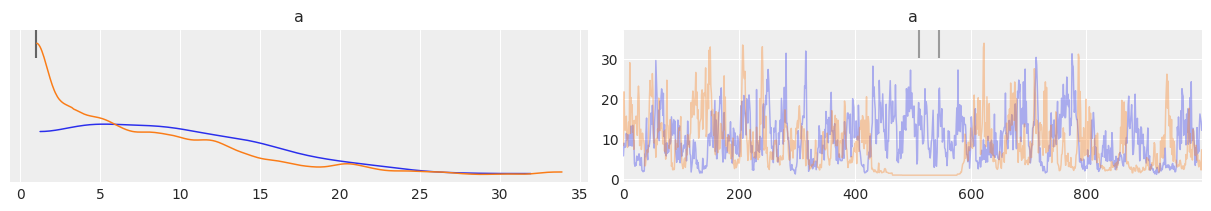
az.plot_trace(trace_cm, var_names=['a'], divergences='top', compact=False)
array([[<AxesSubplot:title={'center':'a'}>,
<AxesSubplot:title={'center':'a'}>]], dtype=object)
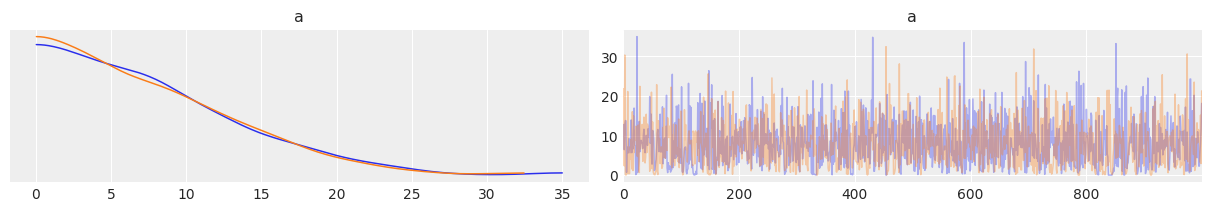
az.plot_trace(trace_ncm, var_names=['a'], compact=False)
array([[<AxesSubplot:title={'center':'a'}>,
<AxesSubplot:title={'center':'a'}>]], dtype=object)
az.summary(trace_cm)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| a | 9.190 | 6.295 | 1.013 | 20.570 | 0.749 | 0.532 | 41.0 | 22.0 | 1.05 |
| b[0] | 0.052 | 11.401 | -24.873 | 20.704 | 0.226 | 0.359 | 2342.0 | 771.0 | 1.02 |
| b[1] | 0.247 | 10.922 | -23.837 | 21.836 | 0.214 | 0.365 | 2497.0 | 936.0 | 1.02 |
| b[2] | 0.039 | 10.862 | -21.110 | 21.709 | 0.216 | 0.362 | 2675.0 | 928.0 | 1.03 |
| b[3] | -0.065 | 11.525 | -22.748 | 23.628 | 0.215 | 0.402 | 2880.0 | 998.0 | 1.02 |
| b[4] | 0.045 | 11.595 | -23.655 | 22.588 | 0.221 | 0.356 | 3088.0 | 825.0 | 1.02 |
| b[5] | 0.340 | 10.935 | -19.502 | 22.569 | 0.217 | 0.399 | 2521.0 | 802.0 | 1.02 |
| b[6] | 0.217 | 12.287 | -26.847 | 23.079 | 0.251 | 0.432 | 2750.0 | 715.0 | 1.03 |
| b[7] | 0.366 | 11.802 | -26.457 | 21.237 | 0.219 | 0.366 | 2806.0 | 996.0 | 1.03 |
| b[8] | 0.381 | 11.643 | -20.940 | 24.580 | 0.226 | 0.338 | 2814.0 | 988.0 | 1.05 |
| b[9] | 0.145 | 10.404 | -19.198 | 21.812 | 0.183 | 0.281 | 3271.0 | 1080.0 | 1.01 |
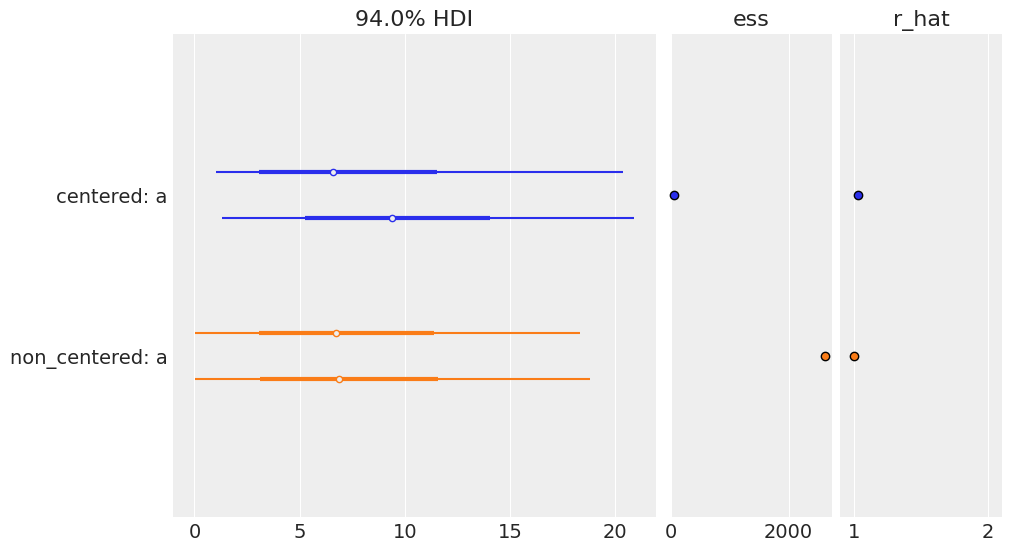
az.rhat(trace_cm)['a'].values, az.rhat(trace_ncm)['a'].values
(array(1.04987792), array(1.00006256))
az.plot_forest([trace_cm, trace_ncm], model_names=['centered', 'non_centered'],
var_names=['a'], r_hat=True, ess=True)
array([<AxesSubplot:title={'center':'94.0% HDI'}>,
<AxesSubplot:title={'center':'ess'}>,
<AxesSubplot:title={'center':'r_hat'}>], dtype=object)
summaries = pd.concat([az.summary(trace_cm, var_names=['a']),
az.summary(trace_ncm, var_names=['a'])])
summaries.index = ['centered', 'non_centered']
summaries
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| centered | 9.190 | 6.295 | 1.013 | 20.570 | 0.749 | 0.532 | 41.0 | 22.0 | 1.05 |
| non_centered | 7.896 | 6.012 | 0.007 | 18.599 | 0.117 | 0.083 | 1567.0 | 727.0 | 1.00 |
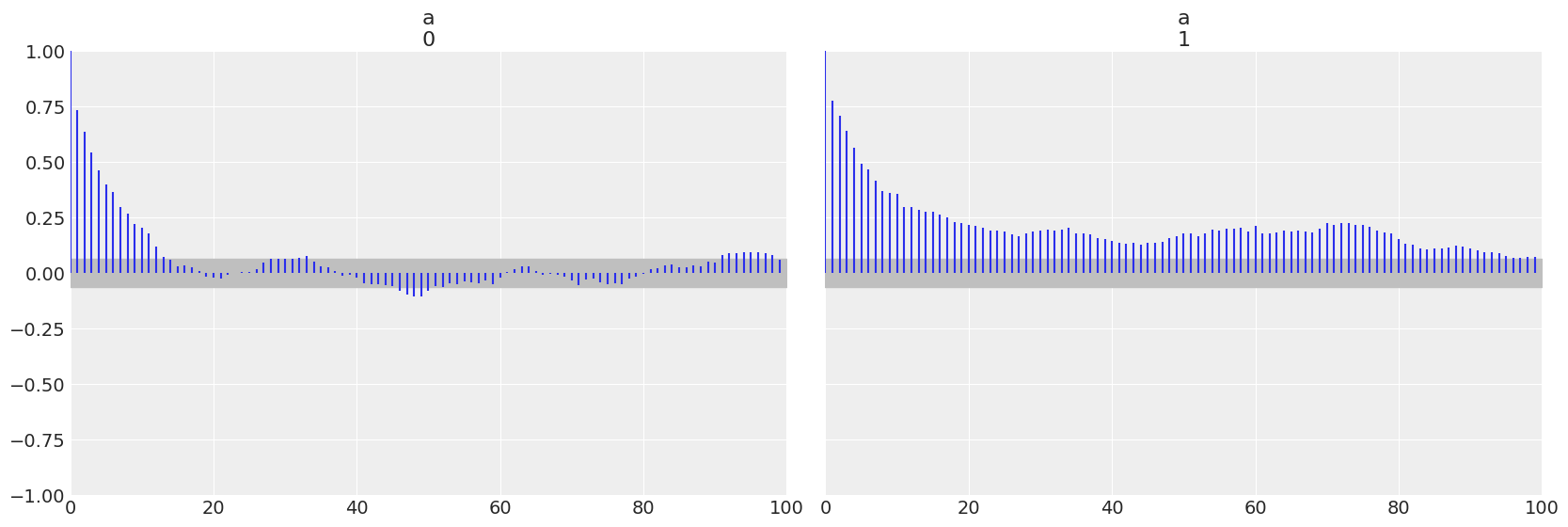
az.plot_autocorr(trace_cm, var_names=['a'])
array([<AxesSubplot:title={'center':'a\n0'}>,
<AxesSubplot:title={'center':'a\n1'}>], dtype=object)
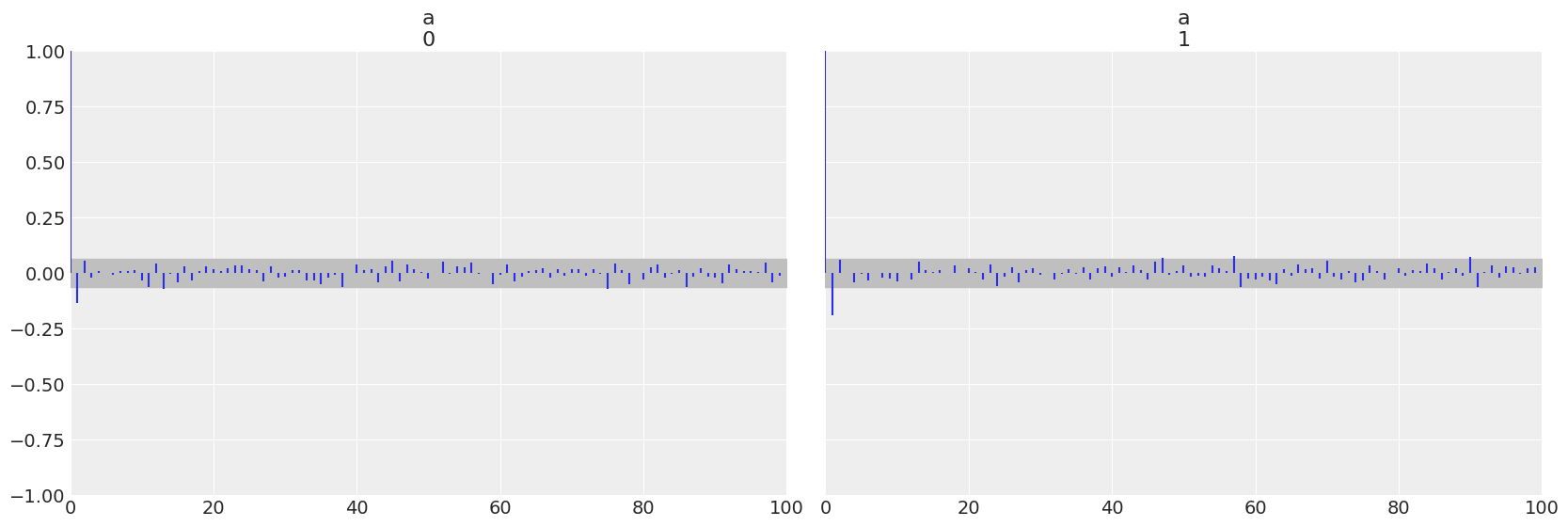
az.plot_autocorr(trace_ncm, var_names=['a'])
array([<AxesSubplot:title={'center':'a\n0'}>,
<AxesSubplot:title={'center':'a\n1'}>], dtype=object)
Effective sample size¶
az.ess(trace_cm)['a'].values
array(41.46579247)
Divergences¶
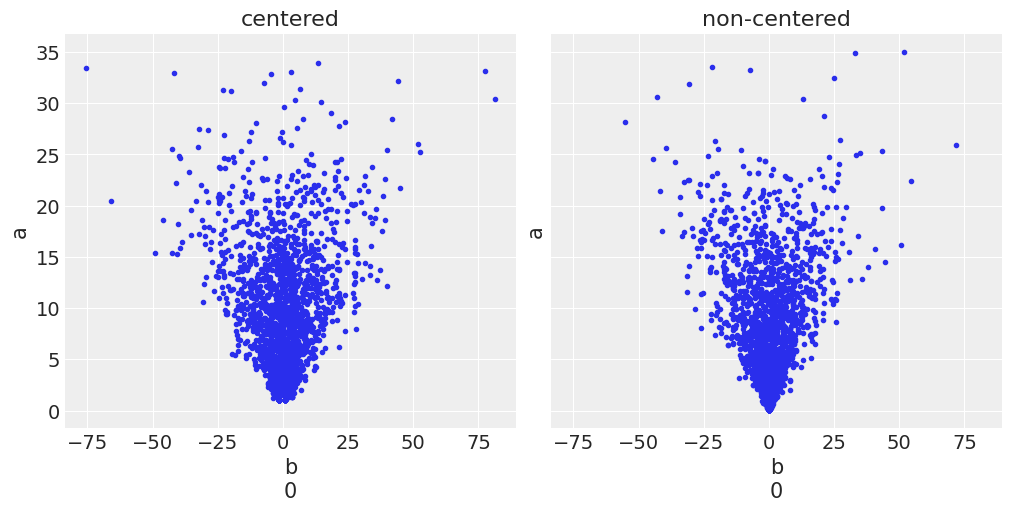
_, ax = plt.subplots(1, 2, sharey=True, sharex=True, figsize=(10, 5), constrained_layout=True)
for idx, tr in enumerate([trace_cm, trace_ncm]):
az.plot_pair(tr, var_names=['b', 'a'], coords={'b_dim_0':[0]}, kind='scatter',
divergences=True, contour=False, divergences_kwargs={'color':'C1'},
ax=ax[idx])
ax[idx].set_title(['centered', 'non-centered'][idx])
UserWarning: Divergences data not found, plotting without divergences. Make sure the sample method provides divergences data and that it is present in the `diverging` field of `sample_stats` or `sample_stats_prior` or set divergences=False
UserWarning: Divergences data not found, plotting without divergences. Make sure the sample method provides divergences data and that it is present in the `diverging` field of `sample_stats` or `sample_stats_prior` or set divergences=False
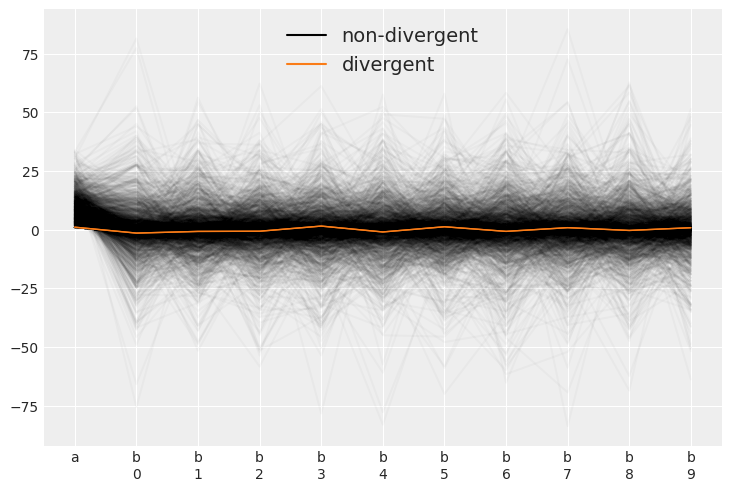
az.plot_parallel(trace_cm)
<AxesSubplot:>