Chapter 6. Mixture Models¶
import os
import warnings
import arviz as az
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import jax.numpy as jnp
from jax import random, vmap, local_device_count, pmap, lax, tree_map
from jax import nn as jnn
from jax.scipy import stats, special
import numpyro
import numpyro.distributions as dist
import numpyro.optim as optim
from numpyro.infer import MCMC, NUTS, HMC, Predictive
from numpyro.diagnostics import hpdi, print_summary
from numpyro.infer import Predictive, SVI, Trace_ELBO, init_to_value
from numpyro.infer.autoguide import AutoLaplaceApproximation
from numpyro.distributions.transforms import OrderedTransform
seed=1234
if "SVG" in os.environ:
%config InlineBackend.figure_formats = ["svg"]
warnings.formatwarning = lambda message, category, *args, **kwargs: "{}: {}\n".format(
category.__name__, message
)
az.style.use("arviz-darkgrid")
numpyro.set_platform("cpu") # or "gpu", "tpu" depending on system
numpyro.set_host_device_count(local_device_count())
# import pymc3 as pm
# import numpy as np
# import scipy.stats as stats
# import pandas as pd
# import theano.tensor as tt
# import matplotlib.pyplot as plt
# import arviz as az
# az.style.use('arviz-darkgrid')
# np.random.seed(42)
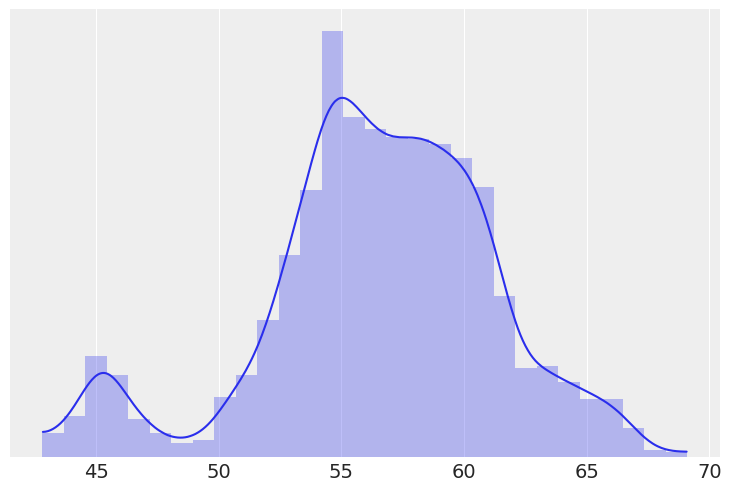
cs = pd.read_csv('../data/chemical_shifts_theo_exp.csv')
cs_exp = cs['exp']
az.plot_kde(cs_exp)
plt.hist(cs_exp, density=True, bins=30, alpha=0.3)
plt.yticks([])
([], [])
#with pm.Model() as model_kg:
# p = pm.Dirichlet('p', a=np.ones(clusters))
# z = pm.Categorical('z', p=p, shape=len(cs_exp))
# means = pm.Normal('means', mu=cs_exp.mean(), sd=10, shape=clusters)
# sd = pm.HalfNormal('sd', sd=10)
#
# y = pm.Normal('y', mu=means[z], sd=sd, observed=cs_exp)
# trace_kg = pm.sample()
# with numpyro.handlers.seed(rng_seed=seed):
# N = 1 # Samples
# b = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(clusters)).expand([N]))
# # dist.Dirichlet(concentration=jnp.ones(clusters)).sample(random.PRNGKey(0), (1,))
# b.squeeze()
clusters = 2
def model(obs=None):
p = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(clusters)))
c = dist.Categorical(probs=p.squeeze())
means = numpyro.sample('means', dist.Normal(loc=cs_exp.mean(), scale=10), sample_shape=(clusters,))
sd = numpyro.sample('sd', dist.HalfNormal(scale=10))
component_dist = dist.Normal(loc=means, scale=sd)
y = numpyro.sample('y', dist.MixtureSameFamily(mixing_distribution=c, component_distribution=component_dist), obs=obs)
kernel = NUTS(model)
mcmc = MCMC(kernel, num_warmup=500, num_samples=2000, num_chains=2, chain_method='sequential')
mcmc.run(random.PRNGKey(seed), obs=jnp.asarray(cs_exp))
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:05<00:00, 491.61it/s, 3 steps of size 6.26e-01. acc. prob=0.92]
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:01<00:00, 1701.92it/s, 7 steps of size 6.96e-01. acc. prob=0.90]
mcmc.get_samples()
{'means': DeviceArray([[46.59215 , 57.53969 ],
[46.77208 , 57.60634 ],
[47.404488, 57.47748 ],
...,
[57.420513, 47.251415],
[57.43893 , 47.13221 ],
[57.534637, 46.69333 ]], dtype=float32),
'p': DeviceArray([[0.07781173, 0.9221883 ],
[0.07711639, 0.9228836 ],
[0.11415413, 0.8858459 ],
...,
[0.90347517, 0.09652483],
[0.8928458 , 0.10715419],
[0.9177794 , 0.08222061]], dtype=float32),
'sd': DeviceArray([3.6385975, 3.6188638, 3.5631063, ..., 3.6311502, 3.6953619,
3.5244987], dtype=float32)}
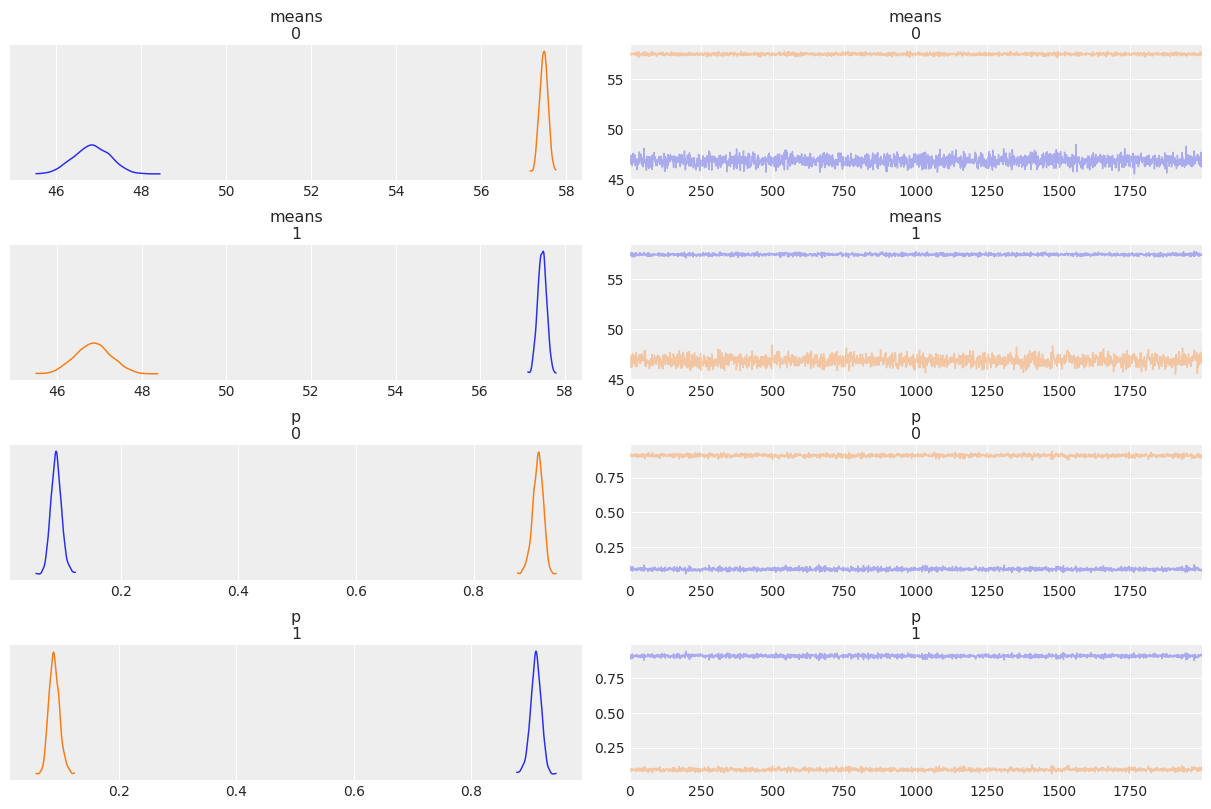
varnames = ['means', 'p']
az.plot_trace(mcmc, varnames, compact=False)
array([[<AxesSubplot:title={'center':'means\n0'}>,
<AxesSubplot:title={'center':'means\n0'}>],
[<AxesSubplot:title={'center':'means\n1'}>,
<AxesSubplot:title={'center':'means\n1'}>],
[<AxesSubplot:title={'center':'p\n0'}>,
<AxesSubplot:title={'center':'p\n0'}>],
[<AxesSubplot:title={'center':'p\n1'}>,
<AxesSubplot:title={'center':'p\n1'}>]], dtype=object)
# clusters = 2
# with pm.Model() as model_mg:
# p = pm.Dirichlet('p', a=np.ones(clusters))
# means = pm.Normal('means', mu=cs_exp.mean(), sd=10, shape=clusters)
# sd = pm.HalfNormal('sd', sd=10)
# y = pm.NormalMixture('y', w=p, mu=means, sd=sd, observed=cs_exp)
# trace_mg = pm.sample(random_seed=123)
# varnames = ['means', 'p']
# az.plot_trace(trace_mg, varnames)
# plt.savefig('B11197_06_06.png')
az.summary(mcmc, var_names=varnames)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| means[0] | 52.154 | 5.324 | 46.283 | 57.672 | 3.752 | 3.176 | 3.0 | 55.0 | 1.83 |
| means[1] | 52.156 | 5.316 | 46.310 | 57.674 | 3.746 | 3.171 | 3.0 | 58.0 | 1.83 |
| p[0] | 0.500 | 0.410 | 0.078 | 0.924 | 0.289 | 0.245 | 3.0 | 51.0 | 1.83 |
| p[1] | 0.500 | 0.410 | 0.076 | 0.922 | 0.289 | 0.245 | 3.0 | 51.0 | 1.83 |
# az.summary(trace_mg, varnames)
Non-identifiability of mixture models¶
jnp.array([.9, 1]) * cs_exp.mean()
DeviceArray([50.859257, 56.51029 ], dtype=float32)
jnp.expand_dims(jnp.asarray(cs_exp), axis=1).shape
(1776, 1)
clusters = 2
def model(obs=None):
p = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(clusters)))
c = dist.Categorical(probs=p.squeeze())
mu = jnp.array([.9, 1]) * cs_exp.mean()
means = numpyro.sample('means', dist.Normal(loc=mu, scale=10), sample_shape=(2,))
sd = numpyro.sample('sd', dist.HalfNormal(scale=10))
component_dist = dist.Normal(loc=means, scale=sd)
y = numpyro.sample('y', dist.MixtureSameFamily(mixing_distribution=c, component_distribution=component_dist), obs=obs)
kernel = NUTS(model)
mcmc2 = MCMC(kernel, num_warmup=500, num_samples=2000, num_chains=2, chain_method='sequential')
mcmc2.run(random.PRNGKey(seed), obs=jnp.expand_dims(jnp.asarray(cs_exp), axis=1))
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:06<00:00, 369.88it/s, 7 steps of size 6.35e-01. acc. prob=0.92]
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:02<00:00, 835.65it/s, 3 steps of size 7.73e-01. acc. prob=0.86]
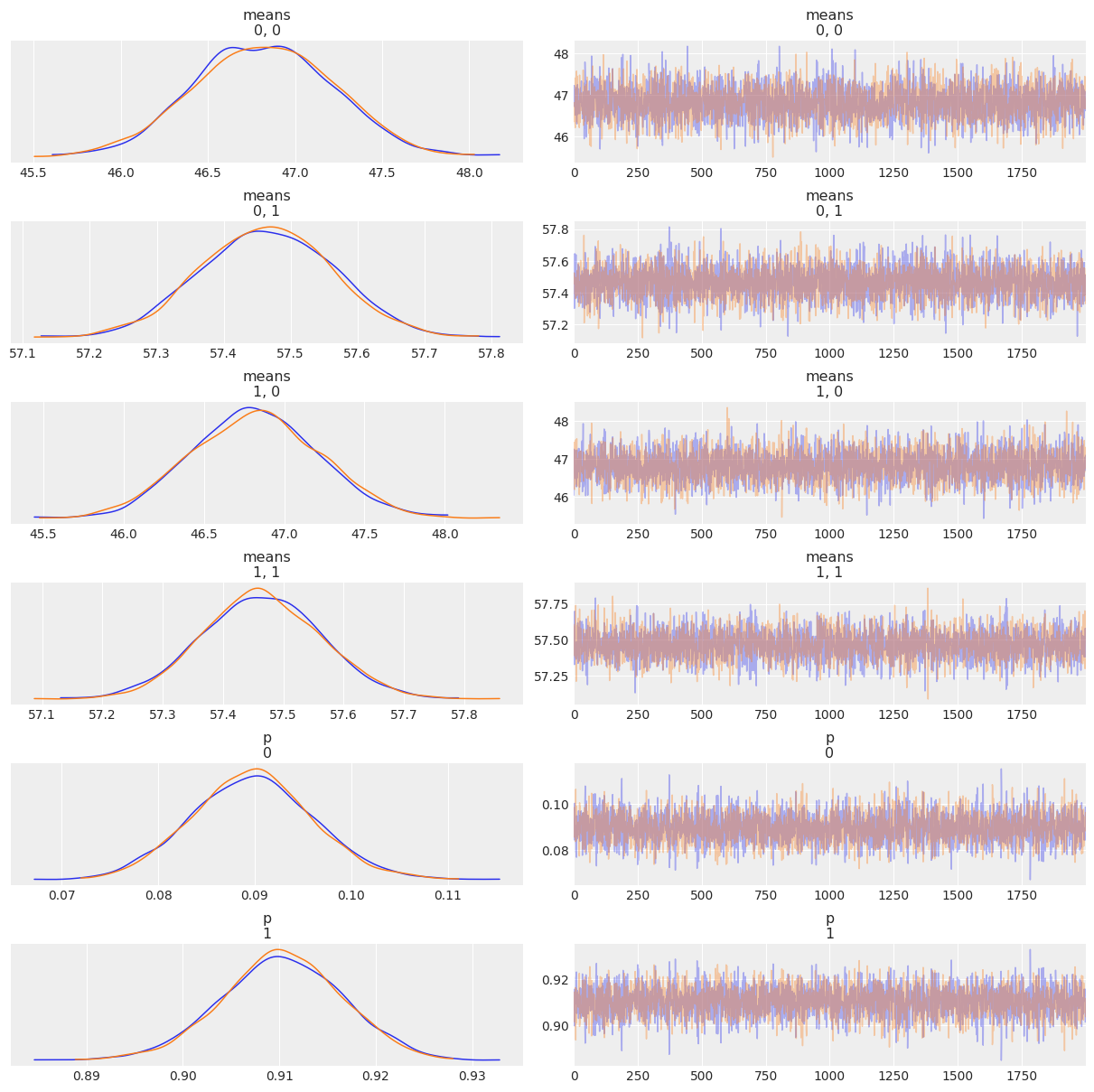
varnames = ['means', 'p']
az.plot_trace(mcmc2, varnames, compact=False)
array([[<AxesSubplot:title={'center':'means\n0, 0'}>,
<AxesSubplot:title={'center':'means\n0, 0'}>],
[<AxesSubplot:title={'center':'means\n0, 1'}>,
<AxesSubplot:title={'center':'means\n0, 1'}>],
[<AxesSubplot:title={'center':'means\n1, 0'}>,
<AxesSubplot:title={'center':'means\n1, 0'}>],
[<AxesSubplot:title={'center':'means\n1, 1'}>,
<AxesSubplot:title={'center':'means\n1, 1'}>],
[<AxesSubplot:title={'center':'p\n0'}>,
<AxesSubplot:title={'center':'p\n0'}>],
[<AxesSubplot:title={'center':'p\n1'}>,
<AxesSubplot:title={'center':'p\n1'}>]], dtype=object)
# instead of a potential we can use an ordered transformation
# transform=pm.distributions.transforms.ordered
az.summary(mcmc2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| means[0,0] | 46.823 | 0.402 | 46.108 | 47.601 | 0.006 | 0.004 | 4707.0 | 3263.0 | 1.0 |
| means[0,1] | 57.464 | 0.099 | 57.274 | 57.646 | 0.001 | 0.001 | 5627.0 | 3227.0 | 1.0 |
| means[1,0] | 46.822 | 0.398 | 46.077 | 47.548 | 0.006 | 0.004 | 4401.0 | 2888.0 | 1.0 |
| means[1,1] | 57.464 | 0.099 | 57.280 | 57.649 | 0.001 | 0.001 | 4593.0 | 3082.0 | 1.0 |
| p[0] | 0.090 | 0.006 | 0.078 | 0.101 | 0.000 | 0.000 | 4095.0 | 2982.0 | 1.0 |
| p[1] | 0.910 | 0.006 | 0.899 | 0.922 | 0.000 | 0.000 | 4095.0 | 2982.0 | 1.0 |
| sd | 3.649 | 0.053 | 3.549 | 3.746 | 0.001 | 0.001 | 5568.0 | 3207.0 | 1.0 |
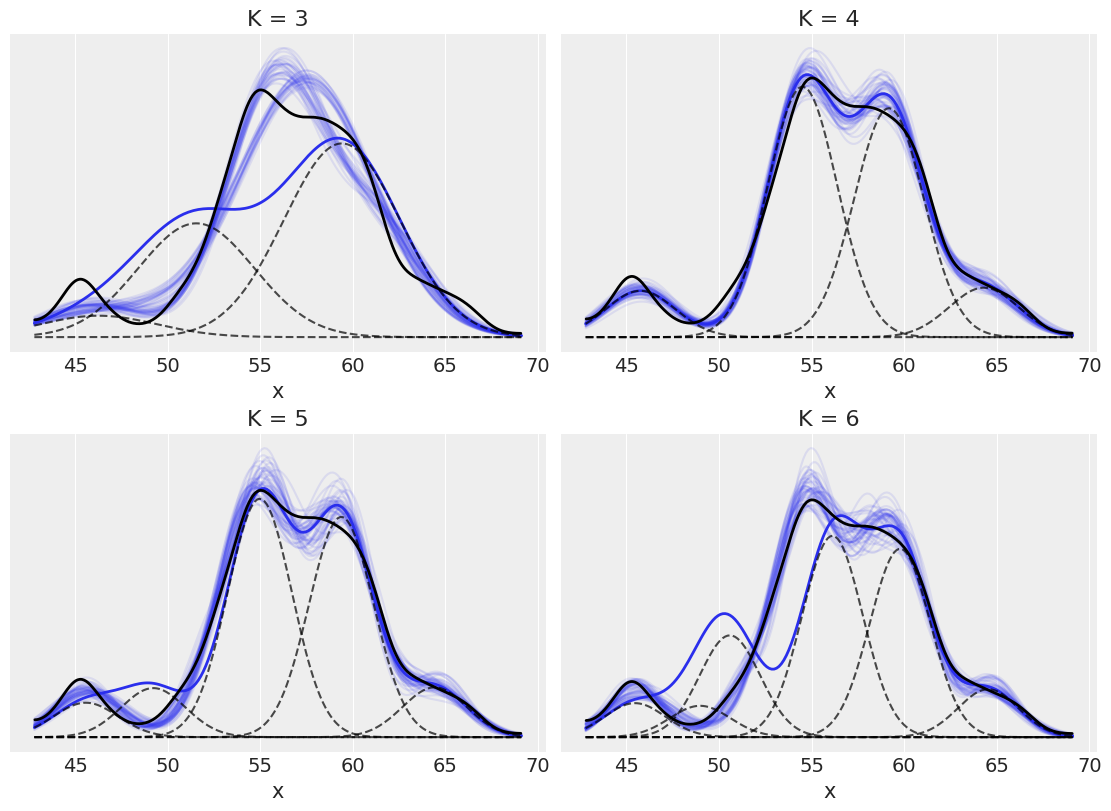
How to choose K¶
def model(obs=None):
p = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(clusters)))
c = dist.Categorical(probs=p.squeeze())
mu = jnp.array([.9, 1]) * cs_exp.mean()
means = numpyro.sample('means', dist.Normal(loc=mu, scale=10), sample_shape=(2,))
sd = numpyro.sample('sd', dist.HalfNormal(scale=10))
component_dist = dist.Normal(loc=means, scale=sd)
y = numpyro.sample('y', dist.MixtureSameFamily(mixing_distribution=c, component_distribution=component_dist), obs=obs)
kernel = NUTS(model)
mcmc2 = MCMC(kernel, num_warmup=500, num_samples=2000, num_chains=2, chain_method='sequential')
mcmc2.run(random.PRNGKey(seed), obs=jnp.expand_dims(jnp.asarray(cs_exp), axis=1))
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:08<00:00, 281.81it/s, 7 steps of size 6.35e-01. acc. prob=0.92]
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:05<00:00, 417.45it/s, 3 steps of size 7.73e-01. acc. prob=0.86]
clusters = [3, 4, 5, 6]
models = []
traces = []
for cluster in clusters:
def model(obs=None):
p = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(cluster)))
c = dist.Categorical(probs=p.squeeze())
mu = jnp.linspace(cs_exp.min(), cs_exp.max(), cluster)
means = numpyro.sample('means',
dist.TransformedDistribution(
base_distribution=dist.Normal(loc=mu, scale=10).expand([cluster]),
transforms=OrderedTransform()
))
print(means)
sd = numpyro.sample('sd', dist.HalfNormal(scale=10))
component_dist = dist.Normal(loc=means, scale=sd)
print(c.probs.shape)
y = numpyro.sample('y', dist.MixtureSameFamily(mixing_distribution=c, component_distribution=component_dist), obs=obs)
kernel = NUTS(model)
trace = MCMC(kernel, num_warmup=500, num_samples=2000, num_chains=2, chain_method='sequential')
trace.run(random.PRNGKey(seed), obs=jnp.expand_dims(jnp.asarray(cs_exp), axis=1))
traces.append(trace)
models.append(model)
[-1.405282 -1.0672494 -0.89249873]
(3,)
Traced<ConcreteArray([-1.405282 -1.0672494 -0.89249873], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([-1.405282 , -1.0672494 , -0.89249873], dtype=float32)
tangent = Traced<ShapedArray(float32[3]):JaxprTrace(level=1/0)>
(3,)
0%| | 0/2500 [00:00<?, ?it/s]
Traced<ShapedArray(float32[3])>with<JVPTrace(level=4/1)>
with primal = Traced<ShapedArray(float32[3])>with<DynamicJaxprTrace(level=2/1)>
tangent = Traced<ShapedArray(float32[3]):JaxprTrace(level=3/1)>
(3,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:08<00:00, 286.52it/s, 63 steps of size 4.23e-02. acc. prob=0.90]
[-1.792481 -0.17571521 0.1223264 ]
(3,)
Traced<ConcreteArray([-1.792481 -0.17571521 0.1223264 ], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([-1.792481 , -0.17571521, 0.1223264 ], dtype=float32)
tangent = Traced<ShapedArray(float32[3]):JaxprTrace(level=1/0)>
(3,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:03<00:00, 820.09it/s, 15 steps of size 2.43e-01. acc. prob=0.91]
[-1.405282 3.7530613 3.927812 4.386582 ]
(4,)
Traced<ConcreteArray([-1.405282 3.7530613 3.927812 4.386582 ], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([-1.405282 , 3.7530613, 3.927812 , 4.386582 ], dtype=float32)
tangent = Traced<ShapedArray(float32[4]):JaxprTrace(level=1/0)>
(4,)
0%| | 0/2500 [00:00<?, ?it/s]
Traced<ShapedArray(float32[4])>with<JVPTrace(level=4/1)>
with primal = Traced<ShapedArray(float32[4])>with<DynamicJaxprTrace(level=2/1)>
tangent = Traced<ShapedArray(float32[4]):JaxprTrace(level=3/1)>
(4,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:35<00:00, 70.17it/s, 255 steps of size 1.19e-02. acc. prob=0.95]
[-1.792481 -0.5164275 -0.2183859 1.5443151]
(4,)
Traced<ConcreteArray([-1.792481 -0.5164275 -0.2183859 1.5443151], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([-1.792481 , -0.5164275, -0.2183859, 1.5443151], dtype=float32)
tangent = Traced<ShapedArray(float32[4]):JaxprTrace(level=1/0)>
(4,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:27<00:00, 90.79it/s, 127 steps of size 8.03e-03. acc. prob=0.94]
[0.7935977 1.8328353 4.7855444 4.9249754 5.1354523]
(5,)
Traced<ConcreteArray([0.7935977 1.8328353 4.7855444 4.9249754 5.1354523], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([0.7935977, 1.8328353, 4.7855444, 4.9249754, 5.1354523], dtype=float32)
tangent = Traced<ShapedArray(float32[5]):JaxprTrace(level=1/0)>
(5,)
0%| | 0/2500 [00:00<?, ?it/s]
Traced<ShapedArray(float32[5])>with<JVPTrace(level=4/1)>
with primal = Traced<ShapedArray(float32[5])>with<DynamicJaxprTrace(level=2/1)>
tangent = Traced<ShapedArray(float32[5]):JaxprTrace(level=3/1)>
(5,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:17<00:00, 145.12it/s, 31 steps of size 1.21e-01. acc. prob=0.95]
[ 0.9015174 4.3690042 7.8917313 8.105168 13.3667755]
(5,)
Traced<ConcreteArray([ 0.9015174 4.3690042 7.8917313 8.105168 13.3667755], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([ 0.9015174, 4.3690042, 7.8917313, 8.105168 , 13.3667755], dtype=float32)
tangent = Traced<ShapedArray(float32[5]):JaxprTrace(level=1/0)>
(5,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:13<00:00, 183.09it/s, 63 steps of size 6.52e-02. acc. prob=0.86]
[0.7935977 1.8328353 7.11053 7.249961 7.460438 8.691059 ]
(6,)
Traced<ConcreteArray([0.7935977 1.8328353 7.11053 7.249961 7.460438 8.691059 ], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([0.7935977, 1.8328353, 7.11053 , 7.249961 , 7.460438 ,
8.691059 ], dtype=float32)
tangent = Traced<ShapedArray(float32[6]):JaxprTrace(level=1/0)>
(6,)
0%| | 0/2500 [00:00<?, ?it/s]
Traced<ShapedArray(float32[6])>with<JVPTrace(level=4/1)>
with primal = Traced<ShapedArray(float32[6])>with<DynamicJaxprTrace(level=2/1)>
tangent = Traced<ShapedArray(float32[6]):JaxprTrace(level=3/1)>
(6,)
sample: 100%|███████████████████████████████████████████| 2500/2500 [00:35<00:00, 70.27it/s, 127 steps of size 6.11e-02. acc. prob=0.57]
[ 0.9015174 4.3690042 5.2567453 5.4701824 10.731791 10.875133 ]
(6,)
Traced<ConcreteArray([ 0.9015174 4.3690042 5.2567453 5.4701824 10.731791 10.875133 ], dtype=float32)>with<JVPTrace(level=2/0)>
with primal = DeviceArray([ 0.9015174, 4.3690042, 5.2567453, 5.4701824, 10.731791 ,
10.875133 ], dtype=float32)
tangent = Traced<ShapedArray(float32[6]):JaxprTrace(level=1/0)>
(6,)
sample: 100%|████████████████████████████████████████████| 2500/2500 [00:25<00:00, 97.44it/s, 31 steps of size 1.19e-01. acc. prob=0.92]
_, ax = plt.subplots(2, 2, figsize=(11, 8), constrained_layout=True)
ax = list(ax.flat)
x = jnp.linspace(cs_exp.min(), cs_exp.max(), 200)
for idx, trace_x in enumerate(traces):
x_ = jnp.array([x] * clusters[idx]).T
for i in range(50):
i_ = int(dist.Uniform(low=0, high=len(trace_x.get_samples()['means'])).sample(key=random.PRNGKey(i)))
means_y = trace_x.get_samples()['means'][i_]
p_y = trace_x.get_samples()['p'][i_]
sd = trace_x.get_samples()['sd'][i_]
distri = dist.Normal(loc=means_y, scale=sd)
ax[idx].plot(x, jnp.sum(jnp.exp(distri.log_prob(x_)) * p_y, 1), 'C0', alpha=0.1)
means_y = trace_x.get_samples()['means'].mean(0)
p_y = trace_x.get_samples()['p'].mean(0)
sd = trace_x.get_samples()['sd'].mean()
distri = dist.Normal(loc=means_y, scale=sd)
#stats.norm(means_y, sd)
ax[idx].plot(x, jnp.sum(jnp.exp(distri.log_prob(x_)) * p_y, 1), 'C0', lw=2)
ax[idx].plot(x, jnp.exp(distri.log_prob(x_)) * p_y, 'k--', alpha=0.7)
az.plot_kde(cs_exp, plot_kwargs={'linewidth':2, 'color':'k'}, ax=ax[idx])
ax[idx].set_title('K = {}'.format(clusters[idx]))
ax[idx].set_yticks([])
ax[idx].set_xlabel('x')
# prior = Predictive(mcmc_l.sampler.model, num_samples=10)
# prior_p = prior(random.PRNGKey(seed), obs=y_1s)
Predictive(model=traces[0].sampler.model,
posterior_samples=traces[0].get_samples(),
return_sites=['y'])(random.PRNGKey(seed)).values()
Traced<ShapedArray(float32[3])>with<DynamicJaxprTrace(level=1/0)>
(3,)
dict_values([DeviceArray([57.74525 , 50.141926, 53.405487, ..., 57.63712 , 58.24072 ,
64.50387 ], dtype=float32)])
ppc_mm = [
Predictive(model=traces[i].sampler.model,
posterior_samples=traces[i].get_samples(),
return_sites=['y'])(random.PRNGKey(seed))
for i in range(4)]
Traced<ShapedArray(float32[3])>with<DynamicJaxprTrace(level=1/0)>
(3,)
Traced<ShapedArray(float32[4])>with<DynamicJaxprTrace(level=1/0)>
(4,)
Traced<ShapedArray(float32[5])>with<DynamicJaxprTrace(level=1/0)>
(5,)
Traced<ShapedArray(float32[6])>with<DynamicJaxprTrace(level=1/0)>
(6,)
type(ppc_mm)
list
for idx, d_sim in enumerate(list(ppc_mm)):
print(idx, d_sim['y'])
0 [57.74525 50.141926 53.405487 ... 57.63712 58.24072 64.50387 ]
1 [55.685463 47.6005 49.555256 ... 57.849586 55.612312 61.546535]
2 [54.532833 46.843277 54.844418 ... 54.875175 53.33296 59.637592]
3 [55.4144 41.105106 53.29952 ... 53.94297 52.922012 60.480656]
jnp.expand_dims(d_sim['y'][:100].T, axis=1).shape
(100, 1)
# iqr(d_sim['y'][:100].T, 0)
# ppc_mm = [pm.sample_posterior_predictive(traces[i], 1000, models[i])
# for i in range(4)]
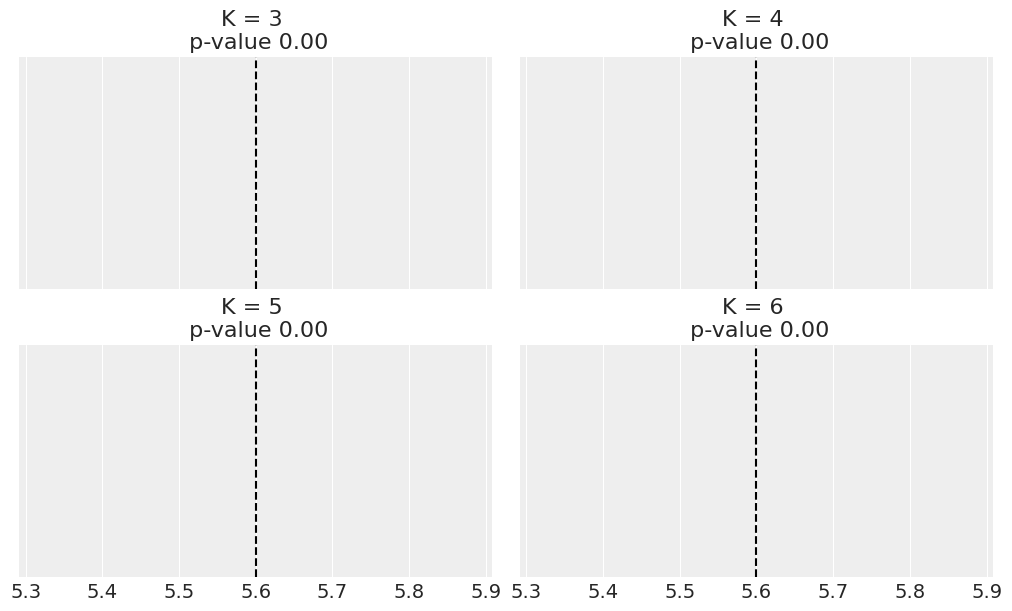
fig, ax = plt.subplots(2, 2, figsize=(10, 6), sharex=True, constrained_layout=True)
ax = list(ax.flat)
def iqr(x, a=0):
return jnp.subtract(*jnp.percentile(jnp.asarray(x), [75, 25], axis=a))
T_obs = iqr(cs_exp)
for idx, d_sim in enumerate(ppc_mm):
ds = jnp.expand_dims(d_sim['y'][:100], axis=1)
T_sim = iqr(ds.T, 0)
print(T_sim)
p_value = jnp.mean(T_sim >= T_obs)
az.plot_kde(T_sim, ax=ax[idx])
ax[idx].axvline(T_obs, 0, 1, color='k', ls='--')
ax[idx].set_title(f'K = {clusters[idx]} \n p-value {p_value:.2f}')
ax[idx].set_yticks([])
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0.]
RuntimeWarning: divide by zero encountered in true_divide
RuntimeWarning: invalid value encountered in true_divide
RuntimeWarning: divide by zero encountered in true_divide
RuntimeWarning: invalid value encountered in true_divide
RuntimeWarning: divide by zero encountered in true_divide
RuntimeWarning: invalid value encountered in true_divide
RuntimeWarning: divide by zero encountered in true_divide
RuntimeWarning: invalid value encountered in true_divide
comp = az.compare(dict(zip(clusters, traces)), ic="waic", method='BB-pseudo-BMA')
comp
UserWarning: The default method used to estimate the weights for each model,has changed from BB-pseudo-BMA to stacking
[5.2118061e+01 7.1559749e+20 8.0297345e+20 8.2045797e+20 2.1260502e+32
1.1329012e+34]
(6,)
Traced<ShapedArray(float32[3])>with<DynamicJaxprTrace(level=1/0)>
(3,)
[5.2118061e+01 7.1559749e+20 8.0297345e+20 8.2045797e+20 2.1260502e+32
1.1329012e+34]
(6,)
Traced<ShapedArray(float32[4])>with<DynamicJaxprTrace(level=1/0)>
(4,)
[5.2118061e+01 7.1559749e+20 8.0297345e+20 8.2045797e+20 2.1260502e+32
1.1329012e+34]
(6,)
Traced<ShapedArray(float32[5])>with<DynamicJaxprTrace(level=1/0)>
(5,)
[5.2118061e+01 7.1559749e+20 8.0297345e+20 8.2045797e+20 2.1260502e+32
1.1329012e+34]
(6,)
Traced<ShapedArray(float32[6])>with<DynamicJaxprTrace(level=1/0)>
(6,)
UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
| rank | waic | p_waic | d_waic | weight | se | dse | warning | waic_scale | |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 0 | -5135.310869 | 15.798141 | 0.000000 | 7.039205e-01 | 32.746687 | 0.000000 | True | log |
| 5 | 1 | -5136.615405 | 12.786784 | 1.304536 | 2.390853e-01 | 31.524217 | 1.300529 | False | log |
| 4 | 2 | -5139.413207 | 7.432724 | 4.102338 | 5.699420e-02 | 31.128302 | 2.552043 | False | log |
| 3 | 3 | -5203.690167 | 17.401520 | 68.379299 | 2.699810e-21 | 31.289554 | 9.751138 | False | log |
# Broken. TODO, fix error relating to this.
# az.plot_compare(comp)
Non-finite mixture model¶
def stick_breaking_truncated(α, H, K):
"""
Truncated stick-breaking process view of a DP
Parameters
----------
α : float
concentration parameter
H : `numpyro` distribution
base distribution
K : int
number of components
Returns
-------
locs : array
locations
w : array
probabilities
"""
# βs = stats.beta.rvs(1, α, size=K)
βs = dist.Beta(concentration1=1, concentration0=α).sample(random.PRNGKey(1), (K,))
w = jnp.empty(K)
w = βs * jnp.concatenate((jnp.array([1.]), jnp.cumprod(1 - βs[:-1])))
locs = H.sample(random.PRNGKey(1), (K,))
return locs, w
# Parameters DP
K = 500
H = dist.Normal()
alphas = [1, 10, 100, 1000]
# plot
_, ax = plt.subplots(2, 2, sharex=True, figsize=(10, 5))
ax = list(ax.flat)
for idx, α in enumerate(alphas):
locs, w = stick_breaking_truncated(α, H, K)
ax[idx].vlines(locs, 0, w, color='C0')
ax[idx].set_title('α = {}'.format(α))
plt.tight_layout()
UserWarning: This figure was using constrained_layout, but that is incompatible with subplots_adjust and/or tight_layout; disabling constrained_layout.
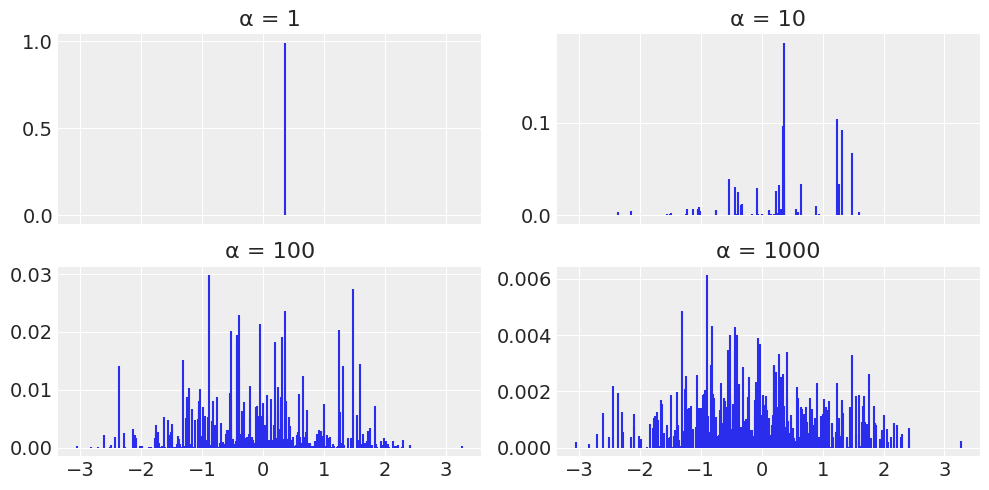
α = 10
H = dist.Normal()
K = 5
x = jnp.linspace(-4, 4, 250)
x_ = jnp.array([x] * K).T
locs, w = stick_breaking_truncated(α, H, K)
# dist = stats.laplace(locs, 0.5)
distri = dist.Laplace(loc=locs, scale=0.5)
plt.plot(x, jnp.sum(jnp.exp(distri.log_prob(x_)) * w, 1), 'C0', lw=2)
plt.plot(x, jnp.exp(distri.log_prob(x_)) * w, 'k--', alpha=0.7)
plt.yticks([])
([], [])
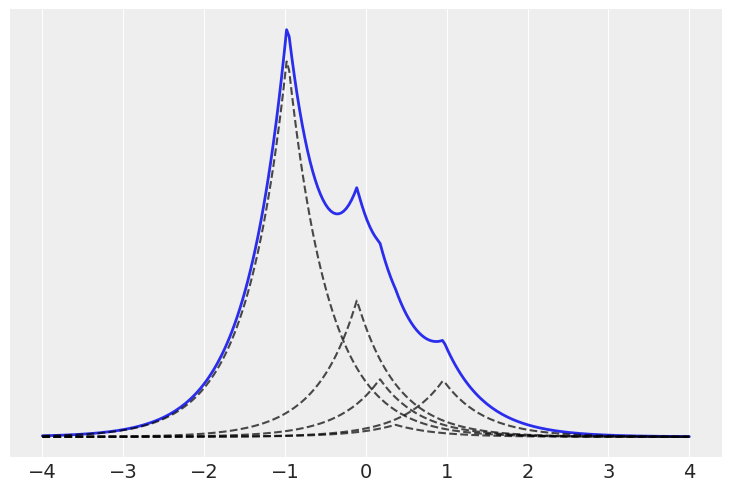
K = 20
def stick_breaking(α, K):
β = numpyro.sample('β', dist.Beta(concentration1=1., concentration0=α), sample_shape=(K,))
w = β * jnp.concatenate([jnp.array([1.]), jnp.cumprod(1. - β)[:-1]])
# β = pm.Beta('β', 1., α, shape=K)
# w = β * pm.math.concatenate([[1.], tt.extra_ops.cumprod(1. - β)[:-1]])
return w
def model(obs=None):
α = numpyro.sample('α', dist.Gamma(concentration=1, rate=1.))
w = numpyro.deterministic('w', stick_breaking(α, K))
# p = numpyro.sample("p", dist.Dirichlet(concentration=jnp.ones(clusters)))
c = dist.Categorical(probs=w.squeeze())
mu = jnp.linspace(cs_exp.min(), cs_exp.max(), K)
means = numpyro.sample('means', dist.Normal(loc=mu, scale=10), sample_shape=(K,))
sd = numpyro.sample('sd', dist.HalfNormal(scale=10))
component_dist = dist.Normal(loc=means, scale=sd)
obss = numpyro.sample('obss', dist.MixtureSameFamily(mixing_distribution=c, component_distribution=component_dist), obs=obs)
kernel = NUTS(model, target_accept_prob=0.85)
mcmc3 = MCMC(kernel, num_warmup=50, num_samples=50, num_chains=2, chain_method='sequential')
mcmc3.run(random.PRNGKey(seed), obs=jnp.expand_dims(jnp.asarray(cs_exp.values), axis=1))
sample: 100%|████████████████████████████████████████████| 100/100 [04:19<00:00, 2.60s/it, 1023 steps of size 6.57e-03. acc. prob=0.52]
sample: 100%|████████████████████████████████████████████| 100/100 [04:46<00:00, 2.86s/it, 1023 steps of size 1.83e-03. acc. prob=0.97]
# with pm.Model() as model:
# α = pm.Gamma('α', 1, 1.)
# w = pm.Deterministic('w', stick_breaking(α, K))
# means = pm.Normal('means',
# mu=np.linspace(cs_exp.min(), cs_exp.max(), K),
# sd=10, shape=K)
# sd = pm.HalfNormal('sd', sd=10, shape=K)
# obs = pm.NormalMixture('obs', w, means, sd=sd, observed=cs_exp.values)
# trace = pm.sample(1000, tune=2000, nuts_kwargs={'target_accept':0.85})
az.plot_trace(mcmc3, var_names=['α'], divergences=False, compact=False);
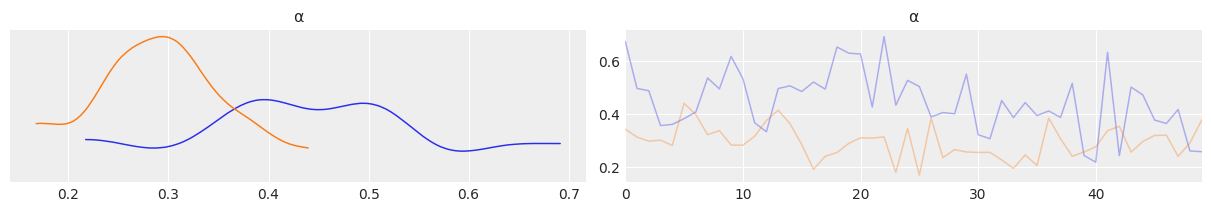
az.plot_trace(mcmc3, var_names=['α'], divergences=False);
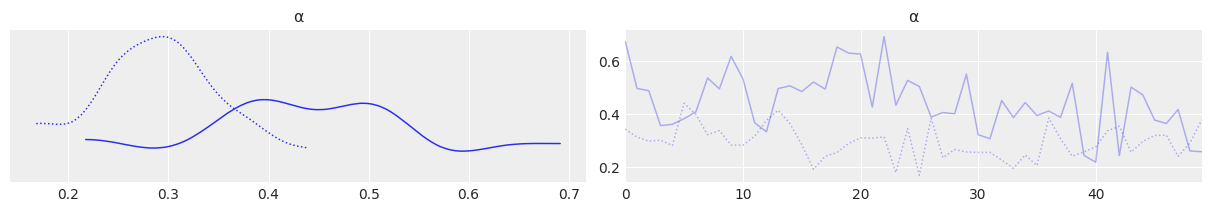
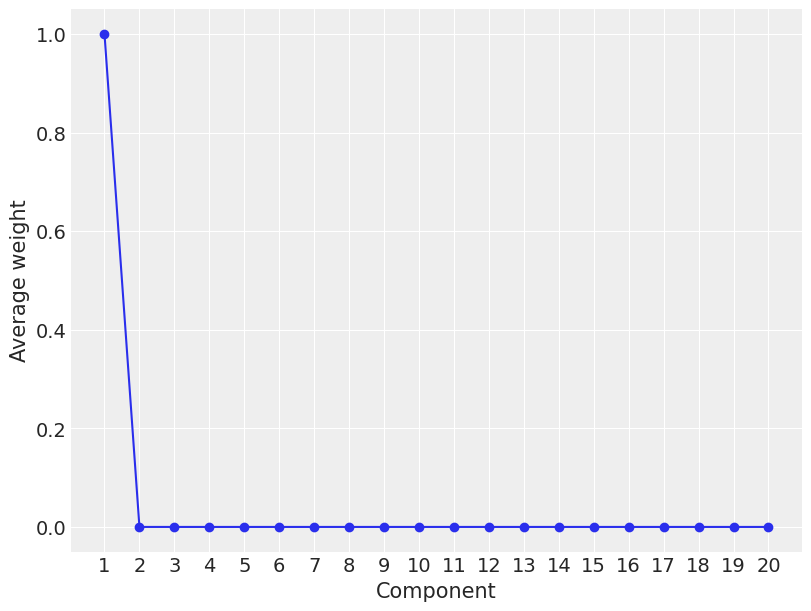
plt.figure(figsize=(8, 6))
plot_w = jnp.arange(K)
plt.plot(plot_w, mcmc3.get_samples()['w'].mean(0), 'o-')
plt.xticks(plot_w, plot_w+1)
plt.xlabel('Component')
plt.ylabel('Average weight')
Text(0, 0.5, 'Average weight')
trace.get_samples()['means']
DeviceArray([[45.43555 , 45.76748 , 45.818993, 54.50315 , 59.173943,
64.33026 ],
[45.743027, 45.879845, 46.071526, 54.450314, 59.080513,
64.30733 ],
[45.39138 , 45.724678, 45.91162 , 54.48335 , 59.17437 ,
64.41409 ],
...,
[45.498985, 51.297474, 54.39981 , 57.54888 , 60.524788,
64.93046 ],
[45.635292, 51.222794, 54.21004 , 57.125477, 60.29111 ,
64.93555 ],
[45.34042 , 51.778076, 54.736847, 57.581596, 60.05674 ,
64.63671 ]], dtype=float32)
mcmc3.get_samples()['means'].shape
(100, 20, 20)
jnp.expand_dims(mcmc3.get_samples()['means'], axis=0).shape
(1, 100, 20, 20)
mcmc3.get_samples()['means'][:, jnp.newaxis, :].shape
(100, 1, 20, 20)
mcmc3.get_samples()['sd'].shape, jnp.expand_dims(mcmc3.get_samples()['sd'], axis=1).shape
((100,), (100, 1))
mcmc3.get_samples()['w'][:, jnp.newaxis, :].shape, jnp.expand_dims(mcmc3.get_samples()['w'], axis=2).shape
((100, 1, 20), (100, 20, 1))
dist.Normal(
loc=jnp.expand_dims(mcmc3.get_samples()['means'], axis=0),
scale=jnp.expand_dims(mcmc3.get_samples()['sd'], axis=0)
)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/var/folders/9y/6kx7fns90pn84gtycx7dyl680000gn/T/ipykernel_33006/463852275.py in <module>
----> 1 dist.Normal(
2 loc=jnp.expand_dims(mcmc3.get_samples()['means'], axis=0),
3 scale=jnp.expand_dims(mcmc3.get_samples()['sd'], axis=0)
4 )
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/distribution.py in __call__(cls, *args, **kwargs)
92 if result is not None:
93 return result
---> 94 return super().__call__(*args, **kwargs)
95
96 @property
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/continuous.py in __init__(self, loc, scale, validate_args)
1238
1239 def __init__(self, loc=0.0, scale=1.0, validate_args=None):
-> 1240 self.loc, self.scale = promote_shapes(loc, scale)
1241 batch_shape = lax.broadcast_shapes(jnp.shape(loc), jnp.shape(scale))
1242 super(Normal, self).__init__(
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/util.py in promote_shapes(shape, *args)
286 else:
287 shapes = [jnp.shape(arg) for arg in args]
--> 288 num_dims = len(lax.broadcast_shapes(shape, *shapes))
289 return [
290 _reshape(arg, (1,) * (num_dims - len(s)) + s) if len(s) < num_dims else arg
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/util.py in wrapper(*args, **kwargs)
193 return f(*args, **kwargs)
194 else:
--> 195 return cached(config._trace_context(), *args, **kwargs)
196
197 wrapper.cache_clear = cached.cache_clear
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/util.py in cached(_, *args, **kwargs)
186 @functools.lru_cache(max_size)
187 def cached(_, *args, **kwargs):
--> 188 return f(*args, **kwargs)
189
190 @functools.wraps(f)
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/lax/lax.py in broadcast_shapes(*shapes)
90 result_shape = _try_broadcast_shapes(shapes)
91 if result_shape is None:
---> 92 raise ValueError("Incompatible shapes for broadcasting: {}"
93 .format(tuple(map(tuple, shapes))))
94 return result_shape
ValueError: Incompatible shapes for broadcasting: ((1, 1, 1, 1), (1, 100, 20, 20), (1, 1, 1, 100))
x_plot = jnp.linspace(cs.exp.min()-1, cs.exp.max()+1, 200)
post_pdf_contribs = jnp.exp(
dist.Normal(
loc=jnp.expand_dims(mcmc3.get_samples()['means'], axis=0),
scale=jnp.expand_dims(mcmc3.get_samples()['sd'], axis=0)
).log_prob(jnp.atleast_3d(x_plot))
) # [:, jnp.newaxis, :]
post_pdfs = (jnp.expand_dims(mcmc3.get_samples()['w'], axis=0) * post_pdf_contribs).sum(axis=-1)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/var/folders/9y/6kx7fns90pn84gtycx7dyl680000gn/T/ipykernel_33006/4200411102.py in <module>
2
3 post_pdf_contribs = jnp.exp(
----> 4 dist.Normal(
5 loc=jnp.expand_dims(mcmc3.get_samples()['means'], axis=0),
6 scale=jnp.expand_dims(mcmc3.get_samples()['sd'], axis=0)
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/distribution.py in __call__(cls, *args, **kwargs)
92 if result is not None:
93 return result
---> 94 return super().__call__(*args, **kwargs)
95
96 @property
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/continuous.py in __init__(self, loc, scale, validate_args)
1238
1239 def __init__(self, loc=0.0, scale=1.0, validate_args=None):
-> 1240 self.loc, self.scale = promote_shapes(loc, scale)
1241 batch_shape = lax.broadcast_shapes(jnp.shape(loc), jnp.shape(scale))
1242 super(Normal, self).__init__(
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/numpyro/distributions/util.py in promote_shapes(shape, *args)
286 else:
287 shapes = [jnp.shape(arg) for arg in args]
--> 288 num_dims = len(lax.broadcast_shapes(shape, *shapes))
289 return [
290 _reshape(arg, (1,) * (num_dims - len(s)) + s) if len(s) < num_dims else arg
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/util.py in wrapper(*args, **kwargs)
193 return f(*args, **kwargs)
194 else:
--> 195 return cached(config._trace_context(), *args, **kwargs)
196
197 wrapper.cache_clear = cached.cache_clear
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/util.py in cached(_, *args, **kwargs)
186 @functools.lru_cache(max_size)
187 def cached(_, *args, **kwargs):
--> 188 return f(*args, **kwargs)
189
190 @functools.wraps(f)
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/lax/lax.py in broadcast_shapes(*shapes)
90 result_shape = _try_broadcast_shapes(shapes)
91 if result_shape is None:
---> 92 raise ValueError("Incompatible shapes for broadcasting: {}"
93 .format(tuple(map(tuple, shapes))))
94 return result_shape
ValueError: Incompatible shapes for broadcasting: ((1, 1, 1, 1), (1, 100, 20, 20), (1, 1, 1, 100))
import numpy as np
x_plot = np.linspace(cs.exp.min()-1, cs.exp.max()+1, 200)
import scipy
post_pdf_contribs = scipy.stats.norm.pdf(np.atleast_3d(x_plot),
mcmc3.get_samples()['means'][:, np.newaxis, :],
mcmc3.get_samples()['sd'][:, np.newaxis, :])
post_pdfs = (mcmc3.get_samples()['w'][:, np.newaxis, :] * post_pdf_contribs).sum(axis=-1)
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
/var/folders/9y/6kx7fns90pn84gtycx7dyl680000gn/T/ipykernel_33006/2613252450.py in <module>
4 post_pdf_contribs = scipy.stats.norm.pdf(np.atleast_3d(x_plot),
5 mcmc3.get_samples()['means'][:, np.newaxis, :],
----> 6 mcmc3.get_samples()['sd'][:, np.newaxis, :])
7 post_pdfs = (mcmc3.get_samples()['w'][:, np.newaxis, :] * post_pdf_contribs).sum(axis=-1)
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/numpy/lax_numpy.py in _rewriting_take(arr, idx, indices_are_sorted, unique_indices)
5150 arr = asarray(arr)
5151 treedef, static_idx, dynamic_idx = _split_index_for_jit(idx, arr.shape)
-> 5152 return _gather(arr, treedef, static_idx, dynamic_idx, indices_are_sorted,
5153 unique_indices)
5154
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/numpy/lax_numpy.py in _gather(arr, treedef, static_idx, dynamic_idx, indices_are_sorted, unique_indices)
5159 unique_indices):
5160 idx = _merge_static_and_dynamic_indices(treedef, static_idx, dynamic_idx)
-> 5161 indexer = _index_to_gather(shape(arr), idx) # shared with _scatter_update
5162 y = arr
5163
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/numpy/lax_numpy.py in _index_to_gather(x_shape, idx, normalize_indices)
5251 def _index_to_gather(x_shape, idx, normalize_indices=True):
5252 # Remove ellipses and add trailing slice(None)s.
-> 5253 idx = _canonicalize_tuple_index(len(x_shape), idx)
5254
5255 # Check for advanced indexing:
/usr/local/anaconda3/envs/bap-numpyro/lib/python3.8/site-packages/jax/_src/numpy/lax_numpy.py in _canonicalize_tuple_index(arr_ndim, idx)
5531 if len_without_none > arr_ndim:
5532 msg = "Too many indices for array: {} non-None/Ellipsis indices for dim {}."
-> 5533 raise IndexError(msg.format(len_without_none, arr_ndim))
5534 ellipses = (i for i, elt in enumerate(idx) if elt is Ellipsis)
5535 ellipsis_index = next(ellipses, None)
IndexError: Too many indices for array: 2 non-None/Ellipsis indices for dim 1.
plt.figure(figsize=(8, 6))
plt.hist(cs_exp.values, bins=25, density=True, alpha=0.5)
plt.plot(x_plot, post_pdfs[::100].T, c='0.5')
plt.plot(x_plot, post_pdfs.mean(axis=0), c='k')
plt.xlabel('x')
plt.yticks([])
## Exercises
# clusters = 3
# n_cluster = [200, 150, 170]
# n_total = sum(n_cluster)
# means = [5, 0, -3]
# std_devs = [2, 2, 2]
# mix = np.random.normal(jnp.repeat(means, n_cluster),
# np.repeat(std_devs, n_cluster))
# az.plot_kde(np.array(mix));
